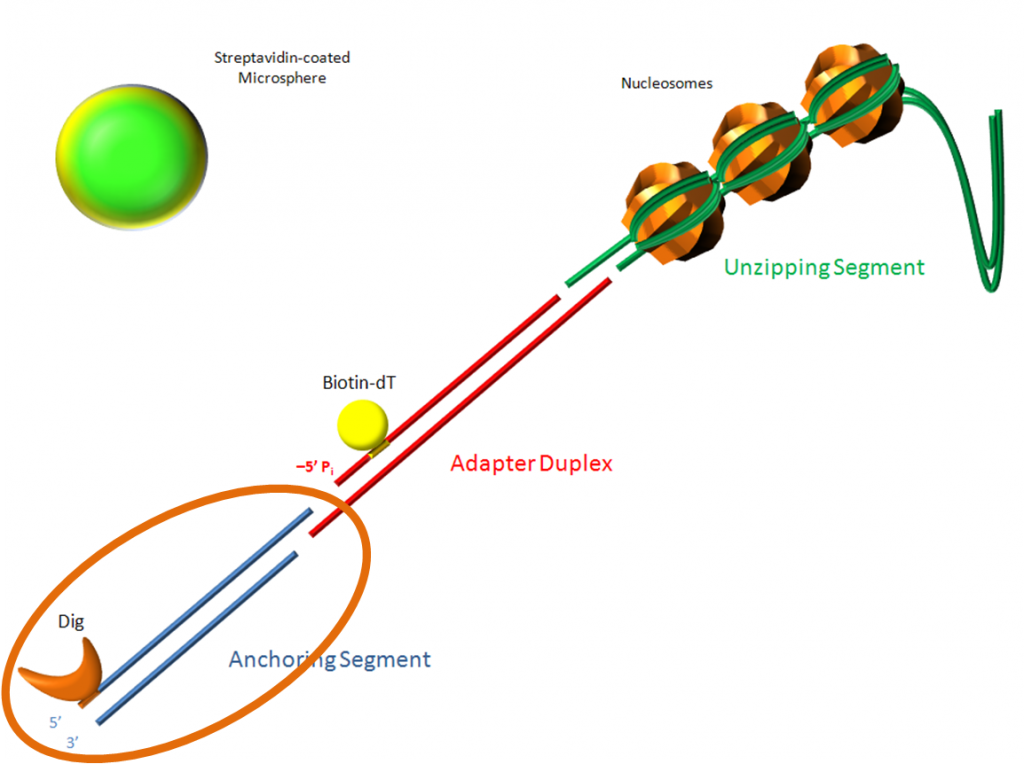
Every time I order adapter sequences, I need to go through the same process. This page lists all the sequences used for the unzipping construct’s adapter duplex. The issue is that I only ever need two sequences: a top and a bottom. The top adapter is easy to pick, but the bottom adapter has two possible solutions and I always forget which one. For this I’ll need to reference some order forms to see what I used last time.
Anyways. The top adapter I need is:
- Top Adapter BstXI/SapI – this adapter has the complementary overhangs for both the BstXI site on the anchor DNA and the SapI site on the unzipping DNA.
And it looks like the bottom adapter I need is the one labeled Bottom Adapter 1a. I’m guessing it is that because: (1) the top adapter on the page is shown annealed to this bottom, and (2) I reference it on this page.
Ok, I’ve verified that the bottom adapter I use most frequently is:
- Bottom Adapter 1a – which I’ve most recently developed two versions of:
- GAGCGGATXACTATACTACATTAGAATTCAGAC – this is the original sequence, and the X is actually a dT-biotin (dT is deoxyribonucleotide thymine)
- TXTXTXAGAGCGGATTACTATACTACATTAGAATTCAGAC – Bottom Adapter 5′ biotin, floppy named because the TXTXTXA is an addition to the 5′ end of the original sequence. The X’s are dT-biotin
- GAGCGGATTACTATACTACATTAGAATTCAGAC – Bottom 5′-biotin adapter named because I’ve removed the dT-biotin and put the biotin at the 5′ end of the sequence.
So unfortunately the verification process I went through is not open. I had to pull my order forms to Alpha DNA to confirm the sequences. Once I found and confirmed the sequences I emailed them to myself. And now I’m posting them here so the entire record is complete. ONS rules!
Anyway, I never remember what Bottom Adapter 1b is for, but I suppose it is not necessary. In the mean time I found a bunch of older notebook entries that contain information about the bottom adapters:
- 11/4/09
- BstXI adapter – I made an adapter to ligate the anchor to itself, and I reference the original bottom adapter called Bottom biotin. I don’t say which one it is, but I’m pretty sure it’s 1a.
I had some other links, but they were either confusing or referred to the bottom adapter without specifying each one. And it is tough searching OWW for the CATG version (1b). Maybe Koch knows? I’ll do some digging later. Research is fun!