Pretty much the same as last time. I’ll need some discussion…
Category Archives: Ligation
Ligation Try 2
Thursday’s ligation failed. So today I’m trying to adjust. Based on Bill Hooker’s suggestion, reducing the amount of ligase could be a solution so I’m trying that. I’m also increasing the ligation time between each adapter addition to give the reduced ligase time to ligate. Finally I’m adding a third reaction using the leftover DNA from Thursday (concentration of EpBR is 60nM and BpALS is 67nM). Check out the reaction below.
Ligation results: FAILED (sort of)
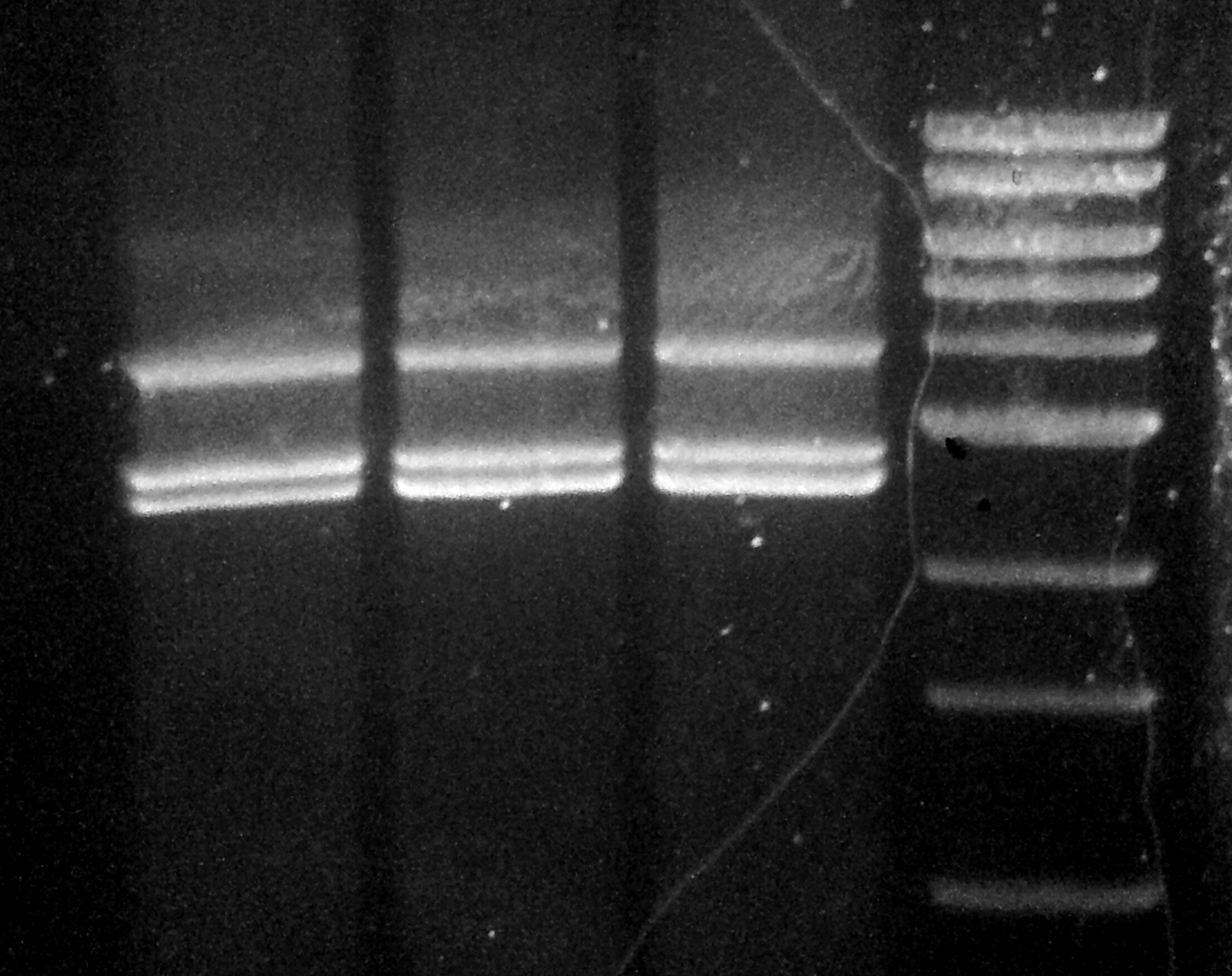
Below you will see the gel of the ligation reaction. It almost worked. I increased the contrast of the image to identify the successful part of the reaction.
The left most and right most lanes are 1kb ladders, and the 4 middle lanes are the ligation reactions. The 1kb ladder weights are (from the top): 10kb, 8, 6, 5, 4, 3, …
At 6.5kb there should be a band, and there is a feint one. There is another band just below that and I have no idea what that could be. The brightest bands are at 4kb (pALS) and 2.5kb (gel extracted pBR322). There is also another band just below that which I suppose is the other fragment of the pBR322 digestion, but I’m not too sure.
Anyways here is the image:
Ligation reaction: pALS-adapter-pBR
I’m almost there! Hopefully in a couple hours I’ll have unzippable DNA. But before we get ahead of ourselves let’s take the final step… together.
I’m setting up two reactions. The first is with some adapter DNA from a couple years ago, that I’ve gotten successful ligation results with. The second is the newer stuff I bought over the summer, that may not have worked all that well (inconclusive). Here is the protocol:
3-piece ligation: EpBR, BpALS, 5′-bio/int-bio adapter results
Lane assignments:
- 1kb ladder
- 5′ bio adapter reaction
- 5′ bio adapter reaction
- int-bio adapter reaction
- int-bio adapter reaction
I had to split each reaction into 2 lanes because the volume of the reaction wouldn’t fit in 1 lane.

So this reaction failed. I’m guessing the digestion of pALS didn’t work. There is an interesting artifact in this gel though. In lanes 2 and 3 you can see 2 bands between 2 and 3 kb. I have no idea why the DNA would separate like that. There are literally only 3 pieces of DNA in here: a ~30bp piece which wouldn’t be visible in this gel, a 2.5kb piece which is the EpBR fragment and is where the 2 bands are, and the 4kb pALS piece which is above these dual bands.
Oh well, I’ll try again at a future date when I run out of the DNA that I have currently produced.
3-piece ligation: BpALS, EpBR, 5′-bio/int-bio adapter
A few days ago I did a 3-piece ligation that didn’t turn out so well. Today’s ligation is more like the traditional ligation that I was supposed to do with the exception that I’m using the pALS anchor instead of the pRL anchor.
I mentioned earlier, that BpALS (pALS anchor digested with BstXI) is better overall because it is longer and makes the optical tweezer operations a little easier and better.
pBR322 can be digested with EarI or SapI for use in our construct for DNA unzipping. I haven’t done any studies that show which method produces better results in terms of final yield, but I’m pretty convinced SapI will because you only have to perform one gel extraction. If you digest pBR with EarI you get two linear fragments. And according to separate analysis by Koch and myself the overhang that we need for the unzipping construct ligation is the longer piece (in the gel image linked above it is the brighter top band).
The benefit to following this method is you can perform a three piece ligation (essentially ligating the entire construct together in one shot) because the EpBR piece (pBR322 digested with EarI) can only ligate to the adapter, and the BpALS/BpRL piece can only ligate to the other end of the adapter (both adapters that I’ve been using work the same way).
And so today I’m doing the proper three-piece ligation reaction. The reaction setup and method can be seen in the table below:
Tetherable DNA concentrations
I finished the gel extraction from the ligations of yesterday. So I decided to spare the 1ul required to measure the concentrations of the DNA for the nanodrop. I also measure the samples from the 3-piece ligations. Here are the resulting concentrations:
- Some nomenclature:
- T#pXX: T stands for tetherable; #indicates which adapter is in the final product (either 5’biotin or internal-biotin); pXX is the plasmid name for the adapter (either pRL or pALS)
- T5pRL (~5.4kb)
- 23.8ng/ul
- 6.6nM
- TIpRL (~5.4kb)
- 9.3ng/ul
- 2.6nM
- T5pRL (~5.4kb, 3-piece result)
- 14.7ng/ul
- 4.1nM
- TIpRL (~5.4kb, 3piece result)
- 50.4ng/ul
- 14nM
- T5pALS (~8.4kb)
- 23.6ng/ul
- 4.3nM
Overall a much better yield than I expected, but I don’t completely trust the nanodrop so take these results to be whatever you want. In my experience I’ve never had good tethering efficiency with anything less than about 100pM DNA so I would recommend dilution of each of these about 1:10 for tethering purposes.
SDM 2-piece ligation results
Here are the lane designations:
- 1kb ladder
- 5’bio reaction from yesterday
- 5’bio reaction from this morning
- internal-bio reaction from yesterday
- internal-bio reaction from this morning


It looks like the ligation didn’t work exactly as planned, but I would say definitely better than the 3 piece ligation debacle. I also let the gel run for too long, so the 1, 1.5, and 2 kb bands have run off the gel. The bottom band in the gel is the 3kb band. The brightest bands in my gel are the 4.3kb linear pBR322 fragment and then what would be the ligation product. And it appears the ligation didn’t work at all in the 5’bio reaction from this morning. Oh well gel extraction and unzipping attempts are next on the “To Do.”
Ligation Part 2
This morning I’m using my leftover supplies from yesterday to do the exact same reaction. All the information needed can be found on that page.
Ligation: 5pBR and IpBR to pRL anchor
After my earlier reaction it is time to continue on. I have purified the ligation reactions from before to remove enzyme and unligated adapter molecules (QiaQuick cleanup kits work really well for purifying short DNA molecules). I ran some samples in the nanodrop to calculate the resulting concentrations (shown in the reaction below, and calculated from the DNA concentration calculator). Next I’m running the reaction below:
- 5pBR – pBR cut with SapI and ligated to the 5′-bio adapter
- IpBR – pBR cut with SapI and ligated to the internal-bio adapter
- pRL anchor – 1.1kb PCR fragment obtained from pRL574 and cut with BstXI