Category Archives: Shotgun DNA Mapping
Defense Outline
Just over a week away now…
- Introduction
- What is D2O?
- The history of D2O
- Gilbert Lewis:
- purification
- biological effects
- The hypothesis
- Joseph Katz
- various experiments
- Gilbert Lewis:
- Uses of D2O
- NMR, mass spec
- The need for a D2O adapted organism
- Experiments in DDW
- use for space travel
- cure for cancer?
- The effects on life
- Tobacco Seeds
- The Crumley experiment and repeating the experiment
- Tobacco seed germination rate
- tobacco seed growth rate in low deuterium concentration
- Arabidopsis
- arabidopsis growth rate
- arabidopsis morphology
- E. coli
- growth rates
- adaptation and adapted growth
- morphology
- Yeast
- growth rates
- adaptation – can’t adapt
- morphology
- stall during cell division
- microtubule stabilization in D2O
- Tobacco Seeds
- Molecular effects
- Stabilization of biomacromolecules
- DLS experiments
- Catalase
- Ovalbumin
- YPD longevity
- DLS experiments
- Investigation of HD exchange
- mechanism and exploitation for protein struture studies
- FT-IR analysis
- Cavity ring-down analysis
- low cost measurement of local atmosphere isotopic composition
- Effect on DNA
- The pursuit of shotgun DNA mapping
- optical tweezers
- methods
- overstretching data
- Stabilization of biomacromolecules
- Future Work
- Arabidopsis
- adaptation
- seed growth in low deuterium
- Tobacco growth in low D2O
- Yeast morphology in taxol
- E coli protein expression in D2O and protein structure analysis
- DNA
- overstretching in D2O with intercalators
- Arabidopsis
Well there is my idea of how to present my dissertation. I’m not sure if/where I should put my discussion on open notebook science. Also there are a couple things that I could see going elsewhere. I could describe the yeast and e. coli stuff in parallel instead of one after another. Also the HD exchange stuff could easily go right after the yeast, e. coli, or even the tobacco seed stuff. What to do…
Otherwise I think the story is pretty compelling: history of D2O and the unanswered question by Lewis. Investigations into D2O effects and trying to understand low D2O concentration effects, effects on macromolecules, and the understanding of large volume/long-term HD exchange.
Any feedback you may have would be GREATLY appreciated. I’ll send you a figshare t-shirt, or if you are XL, I’ll send you a hoodie (but I only have one).
Ligation try 2 results
Ligation Try 2
Thursday’s ligation failed. So today I’m trying to adjust. Based on Bill Hooker’s suggestion, reducing the amount of ligase could be a solution so I’m trying that. I’m also increasing the ligation time between each adapter addition to give the reduced ligase time to ligate. Finally I’m adding a third reaction using the leftover DNA from Thursday (concentration of EpBR is 60nM and BpALS is 67nM). Check out the reaction below.
Ligation results: FAILED (sort of)
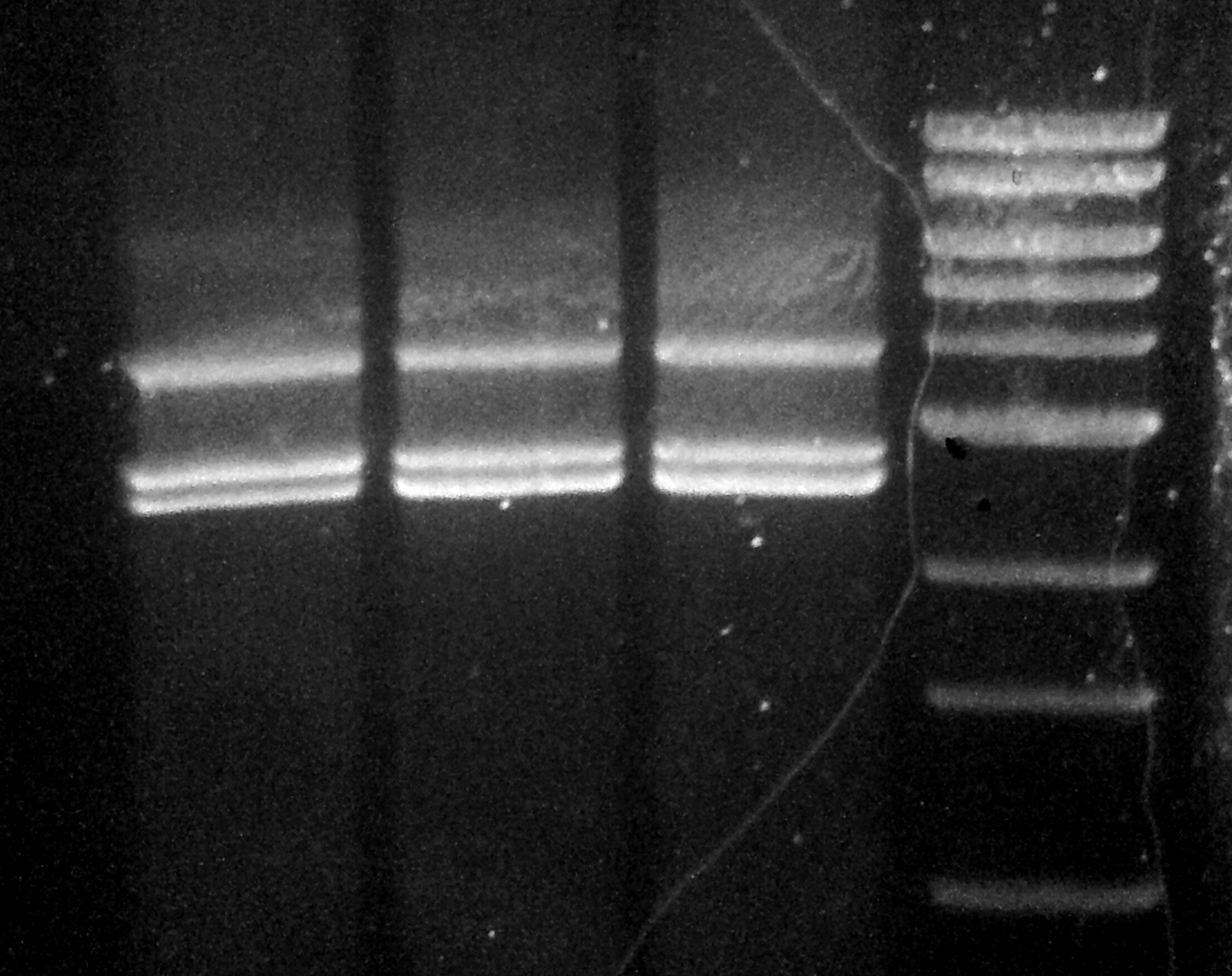
Below you will see the gel of the ligation reaction. It almost worked. I increased the contrast of the image to identify the successful part of the reaction.
The left most and right most lanes are 1kb ladders, and the 4 middle lanes are the ligation reactions. The 1kb ladder weights are (from the top): 10kb, 8, 6, 5, 4, 3, …
At 6.5kb there should be a band, and there is a feint one. There is another band just below that and I have no idea what that could be. The brightest bands are at 4kb (pALS) and 2.5kb (gel extracted pBR322). There is also another band just below that which I suppose is the other fragment of the pBR322 digestion, but I’m not too sure.
Anyways here is the image:
Ligation reaction: pALS-adapter-pBR
I’m almost there! Hopefully in a couple hours I’ll have unzippable DNA. But before we get ahead of ourselves let’s take the final step… together.
I’m setting up two reactions. The first is with some adapter DNA from a couple years ago, that I’ve gotten successful ligation results with. The second is the newer stuff I bought over the summer, that may not have worked all that well (inconclusive). Here is the protocol:
pBR322 gel results and extraction
Yay, good digestion! Here is some useful information about which piece to gel extract. I’m always looking for this mostly for doubling checking purposes, but I need to figure out a place to store this so I can EASILY find it. For now all that matters is I cut out the larger band (top) and do gel cleanup. Onward and upward I suppose…
I extracted into 2 tubes and here are the final amounts:
- 88.7ng/ul –> 52nM
- 101.9ng/ul –> 60nM
pALS BstXI digestion results
The digestion is complete. There is really no way for me to verify the results so I just went ahead and did an enzyme cleanup and purified the DNA. Now I have:
- Tube 1 – 170.0ng/ul –> 63.75 nM
- Tube 2 – 178.3ng/ul –> 66.86nM
Once I verify the pBR322 digestion and gel extract (coming up next) I can set up the ligation reaction and finish up!
PCR yield and BstXI digestion
Since all the reactions yesterday worked, I purified the DNA and took some measurements in the nanodrop:
- Tube 1: 289.6ng/ul in 30ul –> 8688ng
- Tube 2: 259.8ng/ul in 30ul –> 7794ng
Since those yields look phenomenal, I decided to run my digestion of the anchor with BstXI. Once I gel extract my pBR322 digestion, I can run the final step, LIGATION! I’ve never completed this process in 2 days, and I hope I didn’t just jinx myself. Anyway, here is the digestion of pALS:
pBR322 Digestion with EarI
This is the DNA that we unzip. First we need to cut it and gel extract it since EarI cuts in two places on the plasmid.