This will probably be the last update. I’m going to (1) need to take a break until after I defend, and (2) need to start a new experiment because the plants are running low on media. I did just buy these awesome 1in diameter test tubes which should give the plants all the media and water they could need for a longer period of time. Anyways let’s go to the pictures:
All posts by Anthony
The value of open research
This post is written to supplement the P2PU Open Science Education Module, and in particular is meant to be an introduction to open research education.
My name is Anthony Salvagno and I’m an open notebook scientist. That basically means that I publish ALL of my research in real-time on the web. All of that research is attributed under a Creative Commons Attribution ShareAlike (CC BY-SA) license, so the information is free for anyone to use.
If the foundation of science is the pursuit of knowledge and to share that knowledge, then why is it acceptable for scientists to hide their research? Why is it ok for publishers to make you pay for that information? Why is it scientific culture to protect data like it is a commodity to be sold?
In truth, that system worked in the past because the technology was limited. Now the technology exists to instill a new culture. But what are the driving forces that would push someone to make this change? Simply put, I was fed up!
I was originally pushed into open science for one simple reason: my advisor was trained in an extremely closed system. In my first year of graduate school (and his lab), I was presented with the concept of open science, which then was barely taking hold. I was surprised to learn that scientific culture wasn’t a naturally open system, and in fact was surprisingly opposed to that concept.
Early in my graduate program, I became frustrated with the way scientific publications were written. I could only understand a small percentage of the articles I was reading, articles that were written by my peers. I knew most graduate students felt the same way. If we are producing the data and writing the papers, then why would we continue to perpetuate the cycle? So I decided that all of my research would be as accessible as possible.
At times, I would need to understand an experimental process, so I would scan the literature and try to repeat experiments to gain a foothold. I became frustrated with the content contained in the methods sections of scholarly work. Often, the methods would be vague, condensed, or just incomplete, and it would cost me time and money trying and failing to repeat experiments. So I began to document my protocols completely, including minor details that could potentially save other scientists a lot of time.
I have also come across scientific results of a questionable nature. Most of the time the results seemed incongruent with my own research, or even just based on my own expertise I knew there was no way to achieve those results. But the scientific process lacked transparency, so there was no way to understand how the researchers obtained their data. So I made sure that my analysis was entirely transparent, and I provide the data during every phase of analysis including the raw data.
In essence, I have become the scientist that I am because of the experiences that I’ve had. Instead of perpetuating the problems that exist in modern scholarly work, I work toward making a change.
I know I’m not the only scientist who has come across the same issues, in addition to other ones. There are a lot of open scientists who work toward the same goals. We hope to bring about a new culture to enhance the speed of science, to improve our collective knowledge, and to make discoveries that would be impossible in the old system. That is why open research practices are important to me, and that is why every scientist should be an open scientist.
Patent law and open notebook science
The speakers gave an example, but the details are fuzzy. They said they would post the slides, so when that happens, I’ll share them and refer to the timeline.
-
Still not sure how #opennotebookscience can be helpful/destructive for patent filings. I’ve never heard so many contradictory rules!
-
Based on the presentation and the discussion afterward, open notebook science can actually be beneficial for patent filings. JC Bradley highlights this below, and even with the law change it seems to be even more useful since first-to-file is the crucial step now.
-
@Thescienceofant here is one way ONS interacts with patent law usefulchem.blogspot.com/201…
-
That was all I had for the presentation. I spoke with the speakers after the presentation, and asked them about the relevance of ONS to this topic. As I mentioned above, the change from first-to-invent to first-to-file makes it a race to patent. The argument is that entities with a lot of money would benefit since they could just patent every idea. The law is made more accessible because first-to-file can be attributed via a provisional application or public disclosure. From that point on, the applicant has 1 year to file a patent.
To me, open notebook science can be a major benefit to the new patent process. Since it does cost money to file a provisional application, ONS (or other web disclosure) would provide a free alternative to the provisional application. The only difference between the two routes is that through ONS, the patent is immediately public information, while the provisional application keeps the idea in secret. Because the patent will eventually be public domain, the incentive to innovate is delayed a bit.
It should be noted that even though ONS makes your idea open, and encourages potential modification it does not encourage stealing of the idea. You are still protected from patent infringement. But if a competitor sees your idea and makes non-trivial (non-obvious) changes to your idea, then they can be granted a new patent. That is no different from how the patent process works anyways.
But to me, filing a provision for every idea you ever come up with and paying $125 every time is a waste of money since you aren’t likely to follow through with every idea. It also gives the US patent office a lot of unnecessary paper work, and could actually stifle innovation and creativity. ONS would in turn allow some one to share their ideas and protect the best ones for the original creator (since you have one year from first disclosure). You could use your resources to fight for the ones you want to keep and allow others to cultivate the ideas that you won’t ever get to work towards.
It was interesting that when I asked the speaker about ONS and patents, while he didn’t say the conflicted (cause they don’t), he did say that it didn’t make sense to pursue both paths. His reasoning was basically what I outlined above, but it also felt like there was a money undertone to it. You can’t make money if you share your ideas. To that I disagree, but that’s an entirely different story altogether. One that I hope to address in April.
Tracking my stress through weight
Since I’ve been told that stress is a factor in weight gain/loss I’m going to openly track my weight over the next few weeks until I defend and submit my dissertation, and potentially after that. Hopefully I can maintain a stable weight or lose a few lbs and avoid gaining weight as I approach the day. Check it out:
YPD aggregation update
Remember this experiment?
Checking in on it, the YPD has already begun it’s aggregation in both cases, but it is more prevalent in the DI YPD case. Which should be expected. At this point I can see a concentration gradient in both solutions, but the full-on aggregation hasn’t happened yet. As is, it’s not worth photographing because there isn’t much difference.
But thinking back to here, I remembered we used a Dynamic Light Scattering Machine (the DynaPro Titan TC by Wyatt), which just uses scattered light to calculate particle size. In the aggregation experiments we didn’t care so much about specific particle size, just that we wanted to see the particles grow bigger.
If in fact the YPD is aggregating, the solution would become cloudier by eye. So since our Nanodrop measures absorbance, we could potentially see the same effect. The catch is that the DLS machine mentioned above measures at 830nm, and my nanodrop measures a broad spectrum from 250nm-700nm. In that range I’m not sure where it is most accurate except at 260nm, 280nm, and 600nm (which is where it measures DNA, proteins, and cell counts).
I’ll do a more thorough study on Monday, but here is the quick analysis:

orange – aggregated DI YPD
green – aggregated D2O YPD
New YPD made
Protocol – with 460ul of Ampicillin added.
The Open IGERT: Review of the Reviews – Grant Declined
Sorry for the weird title, I wanted to describe as much about this post as possible in the title without making it super huge. Long story short, the IGERT Grant that my peers and I submitted in August has been reviewed and the reviews are in.
The Open IGERT has been declined.
I’m not at all surprised. I knew we were a long shot. And I knew that we weren’t focusing on issues that, while not required or mentioned by the NSF, would be things they would want us to focus on. But, I thought we put together a powerful program. Unfortunately the NSF and their army of anonymous peer reviewers thought otherwise.
So in an effort to improve upon the program I am going to share the reviews with you all, and comment on the reviews. The reviews and the review summary can be found in my Google Drive Folder. Feel free to poke around. For reference, here is the original NSF call for proposals. And so you can skip the link, here is the main objectives of the IGERT call:
- …NSF recognizes the need to educate and support a next generation of researchers able to address fundamental challenges in
- core techniques and technologies for advancing big data science and engineering;
- analyzing and dealing with challenging computational and data enabled science and engineering (CDS&E) problems, and
- researching, providing, and using the cyberinfrastructure that makes cutting-edge CDS&E research possible in any and all disciplines.
On to the reviews:
Continue reading The Open IGERT: Review of the Reviews – Grant Declined
Arabidopsis Update
Here is the update for my arabidopsis growth. It still seems to me that the plants in 10% D2O are doing better than the plants in DDW. I’ll have to make a huge purchase of D2O and DDW (still don’t understand why DDW is more expensive than D2O since one is a by product of the other) and try lower concentrations of deuterium (like 0-10% amounts).
Notice that the seeds in 99% D2O germinated (mostly likely due to D-exchange), but haven’t grown in 3 weeks.
I wanted to take images of the root growth, but my camera couldn’t see clearly enough in the agar to take decent pictures.
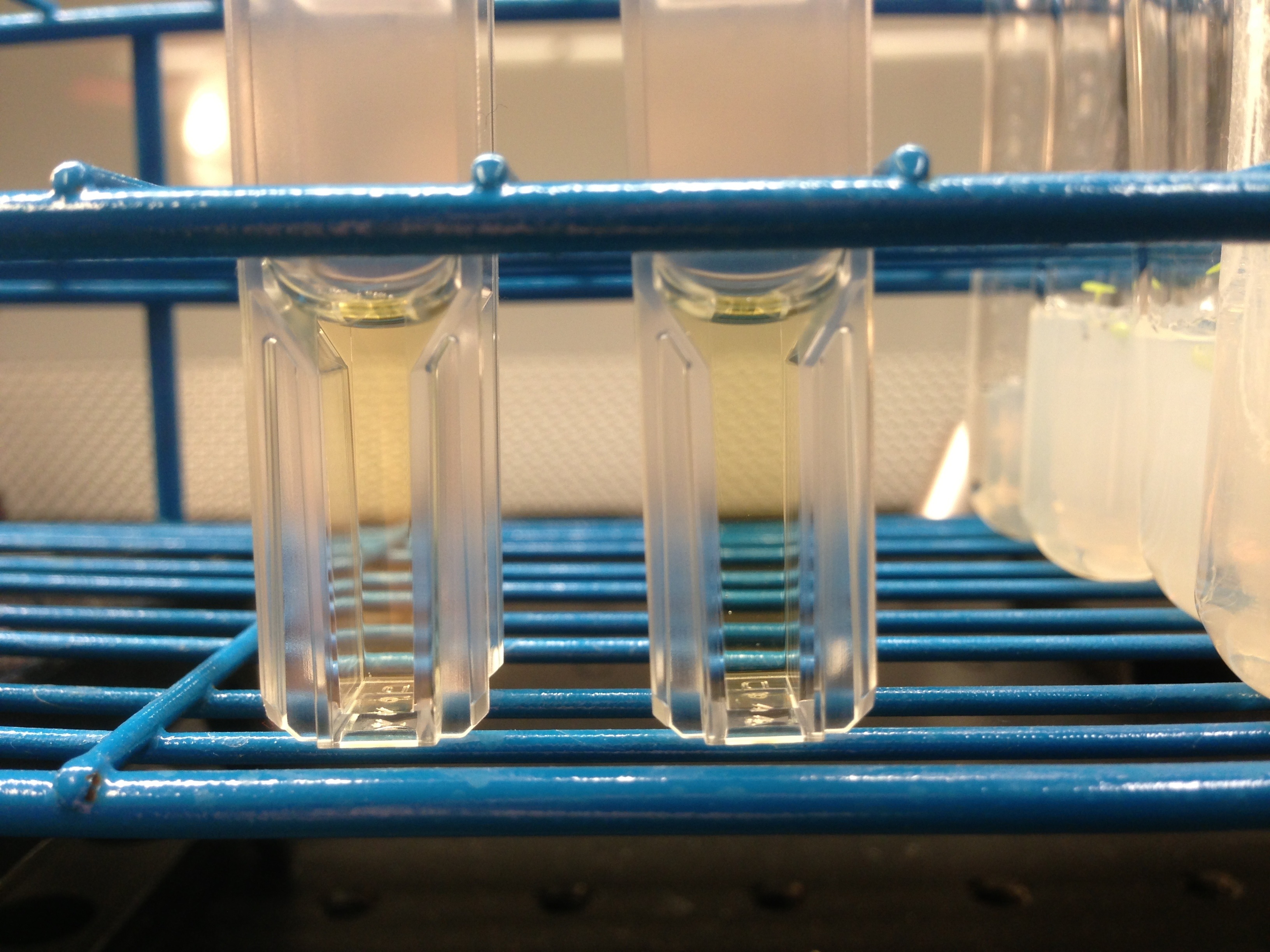
YPD aggregation study
Two summers ago Kenji Doering (an REU student that summer) and I did some protein aggregation studies comparing D2O and DI water. The results were pretty consistent in that proteins don’t aggregate under the same conditions in both samples. What that meant was the D2O is almost certainly better for longevity of chemicals.
A few months ago I noticed that my ypd stocks go noticeably bad after a couple of weeks. But they seemed to last longer in D2O, I just never quantified that. Well now I’m going to.
I have two cuvettes sitting on my bench. One is filled with 1ml of DI YPD and the other is 1ml of D2O YPD. They are sitting next to the benchtop cooler to supply a bit of heat to speed up the aggregation reaction. I will take daily pictures of the solutions to compare the aggregation times of each media. Here is the day=0 time point (on the left is the DI YPD sample, and the right is the 99% D2O sample):
Arabidopsis Update: From Feb 13, 2013 (6 days ago)
These pictures are a bit late, but better late than never. They are an update of the growth progress of arabidopsis in varying amounts of D2O. The group shot is inconsistent with the others because the sample captured for 0% D2O (DDW, deuterium depleted water) is different than the sample used in the individual 0% D2O image.
With that said, there are a couple morphological things I would like to point out. In my opinion the plants are growing better (bigger, faster, etc) in 10% D2O than they are in DDW. They also appear to be slightly more green. And finally there are little hairs on the leaves that seem to be more prominent than in the other samples.
Meanwhile, in 60% D2O the plants are very yellow in appearance. It is interesting that more plants sprouted in that sample, but they are significantly behind in development.
I’ll update tomorrow for the weekly update. And this time I’ll upload the images in real-time. Promise!