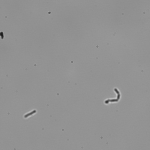
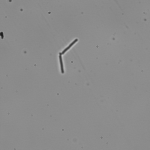
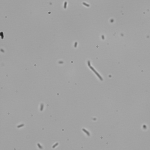
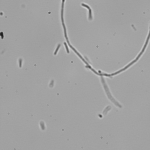
This article is actually the introduction to my dissertation and I thought I’d share it with the world officially rather than let it die in an electronic archive somewhere. I’ve shared this story in some form or another several times already, but I’ve never provided the entire account like this. And so, it is with great pleasure that I share with you, the story of how I became the scientist that I am today…
I joined the KochLab in the Spring of 2007. It was a brand new lab that, at the time, was comprised of Dr. Koch, myself, and my best friend Larry Herskowitz (who is now Dr. Herskowitz). In our first lab meeting, Dr. Koch discussed his scientific endeavors up to that point (some of which are continued in this dissertation) and introduced the concept of open science.
Open science was, and still is, an emerging paradigm, and is not to be confused with a particular field of science. The core concept of open science is providing access information and it is through the opening of scientific research that many new endeavors have become possible. Many of these endeavors have changed the way scientists approach research and acquire data. Citizen science, for instance, has brought a mass scale of human analysis to previously unsolvable problems. Even sharing data has led to new forms of collaboration. Data repositories have allowed scientists to share data with the world in hopes of finding new uses for the shared data. Tools like DataOne have emerged to provide some organization to the new data. Meanwhile, open notebook science has emerged to open the entire scientific process and practitioners make every stage of research accessible including protocols, raw data, data analysis, and much more open to scrutiny.
Continue reading The entire story of my scientific career →
Like this:
Like Loading...